Reconciling Membrane Protein Simulations with Experimental DEER

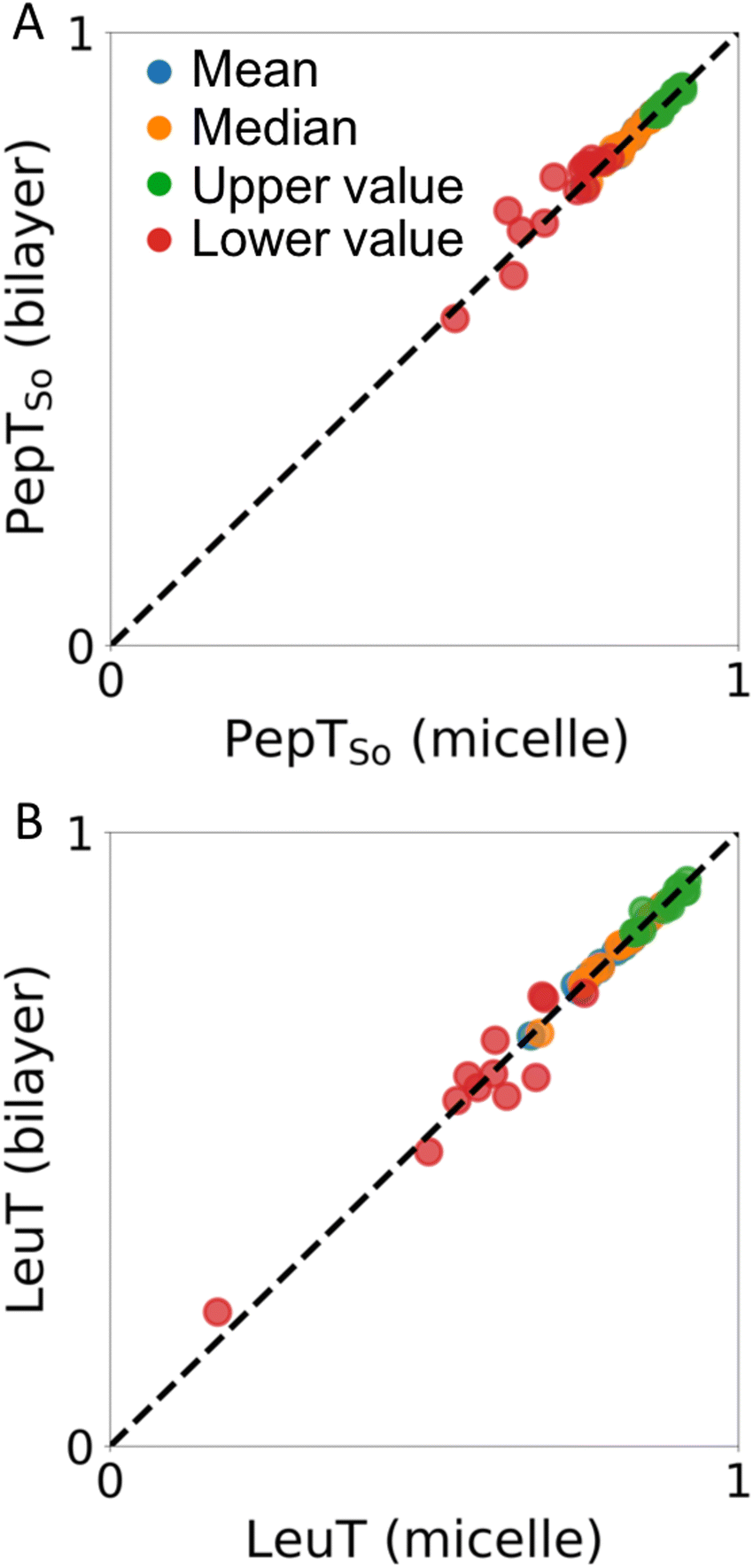
A) Cross-validated scores for 4300 possible DEER residue-pair sets.

Cross-linking, DEER-spectroscopy and molecular dynamics confirm the inward facing state of P-glycoprotein in a lipid membrane - ScienceDirect

Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules

Cross-linking, DEER-spectroscopy and molecular dynamics confirm the inward facing state of P-glycoprotein in a lipid membrane - ScienceDirect
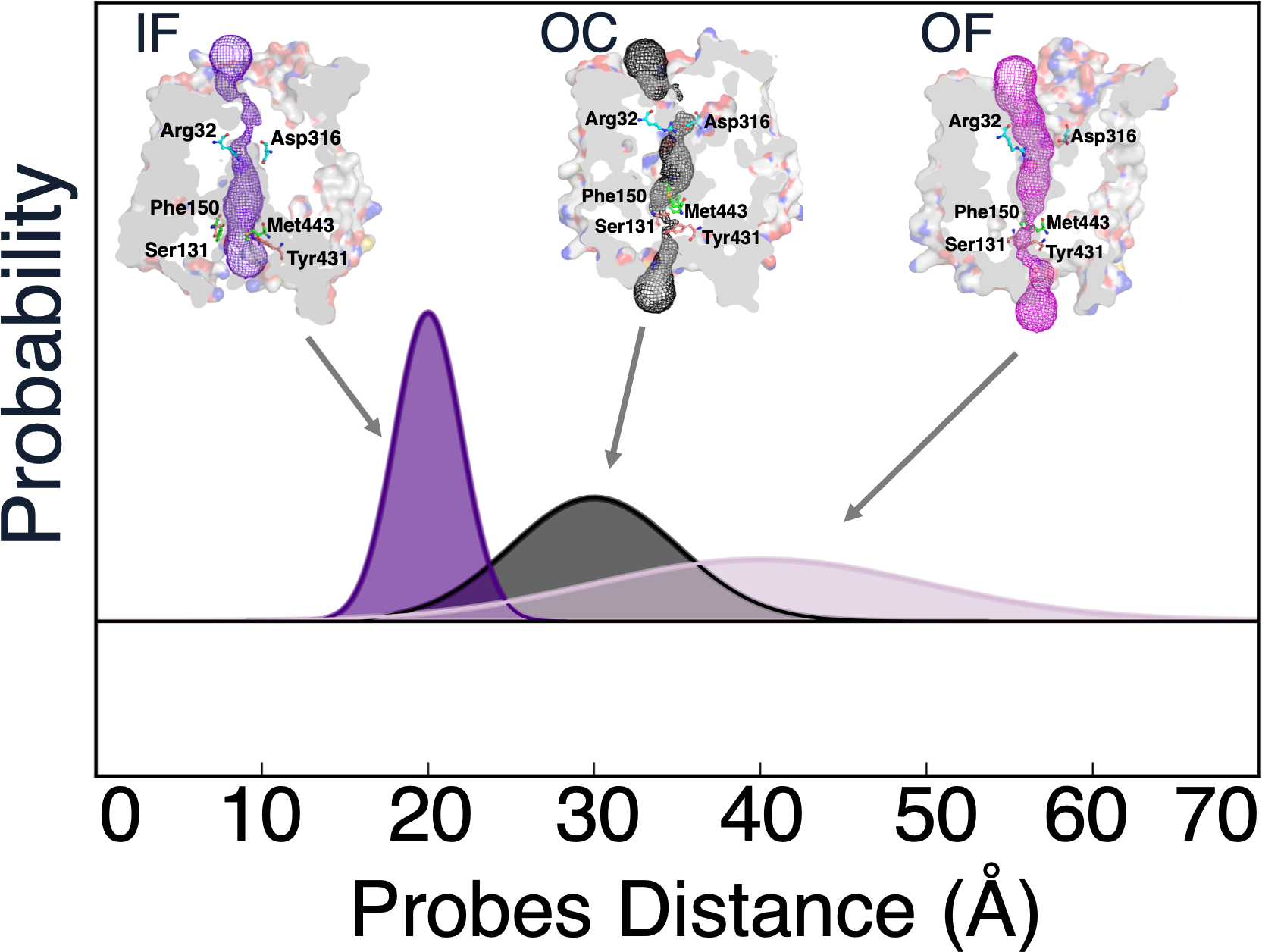
Reconciling membrane protein simulations with experimental DEER spectroscopy data - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/D2CP02890E

DEER Spectroscopy Measurements Reveal Multiple Conformations of HIV-1 SOSIP Envelopes that Show Similarities with Envelopes on Native Virions - ScienceDirect

PDF) Reconciling Membrane Protein Simulations with Experimental DEER Spectroscopy Data

The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells

PDF) Reconciling Membrane Protein Simulations with Experimental DEER Spectroscopy Data

In situ observation of conformational dynamics and protein ligand–substrate interactions in outer-membrane proteins with DEER/PELDOR spectroscopy

DEER EPR Measurements for Membrane Protein Structures via Bifunctional Spin Labels and Lipodisq Nanoparticles

Methods – Shukla Group

A facile approach for the in vitro assembly of multimeric membrane transport proteins