Molecular recognition of trehalose and trehalose analogues by Mycobacterium tuberculosis LpqY-SugABC


Trehalose-containing glycolipids from M. tuberculosis. LOS

Ribbon model of TMBP with bound trehalose (shown as a ball and stick

A Fluorogenic Trehalose Probe for Tracking Phagocytosed Mycobacterium tuberculosis. - Abstract - Europe PMC

Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter - ScienceDirect
PDF) Trehalose Recycling Promotes Energy-Efficient Biosynthesis of the Mycobacterial Cell Envelope

Chemical probes for tagging mycobacterial lipids - ScienceDirect

Structure of the trehalose transporter in the catalytic intermediate

Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter - ScienceDirect
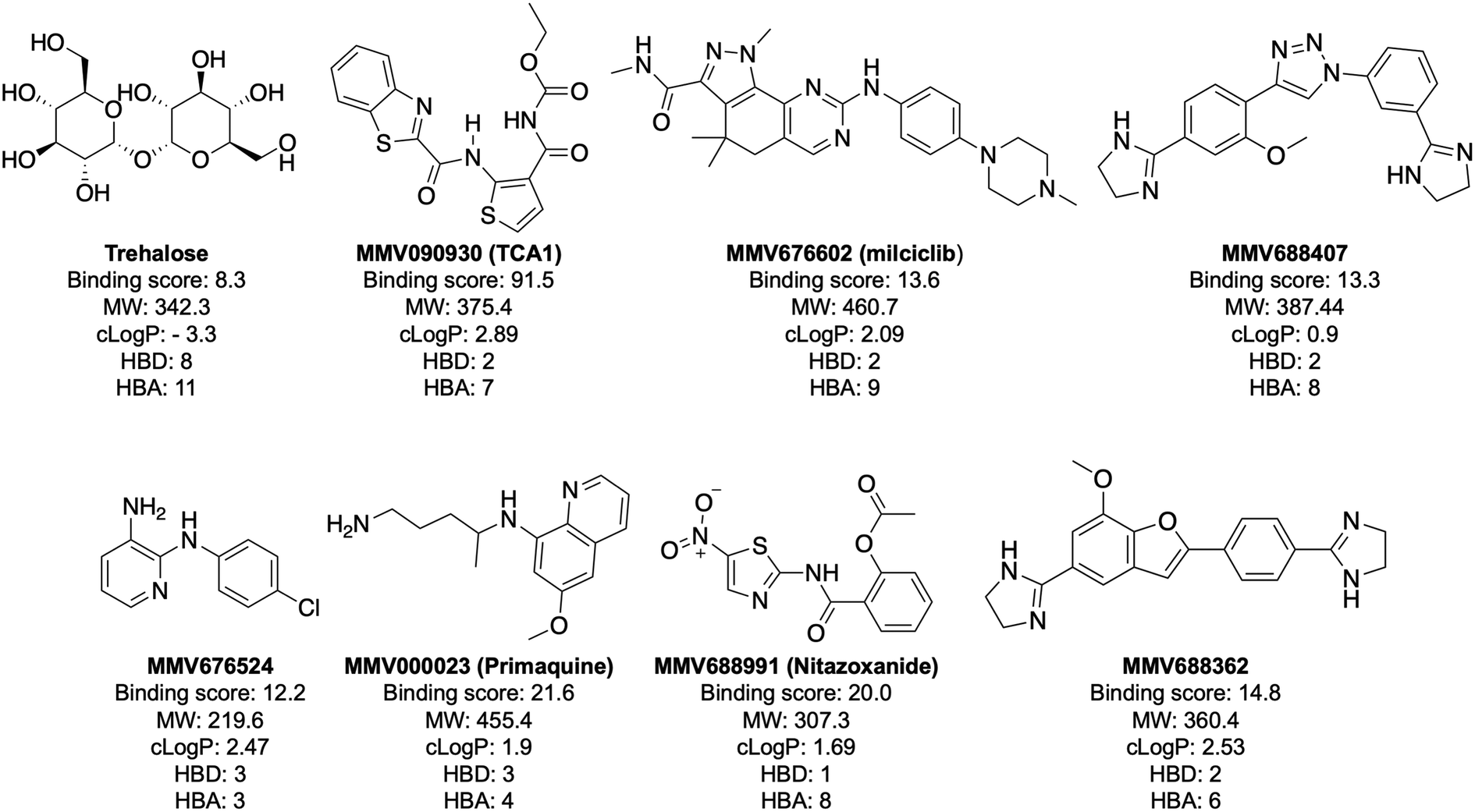
Interrogation of the Pathogen Box reveals small molecule ligands against the mycobacterial trehalose transporter LpqY-SugABC - RSC Medicinal Chemistry (RSC Publishing) DOI:10.1039/D2MD00104G

Trehalose is incorporated into M. tuberculosis in vitro and in

Schematic representation of the M. tuberculosis cell envelope and

Trehalose binding site in Mtr LpqY. A, illustration of the Mtr LpqY

Probing the Mycobacterial Trehalome with Bioorthogonal Chemistry









