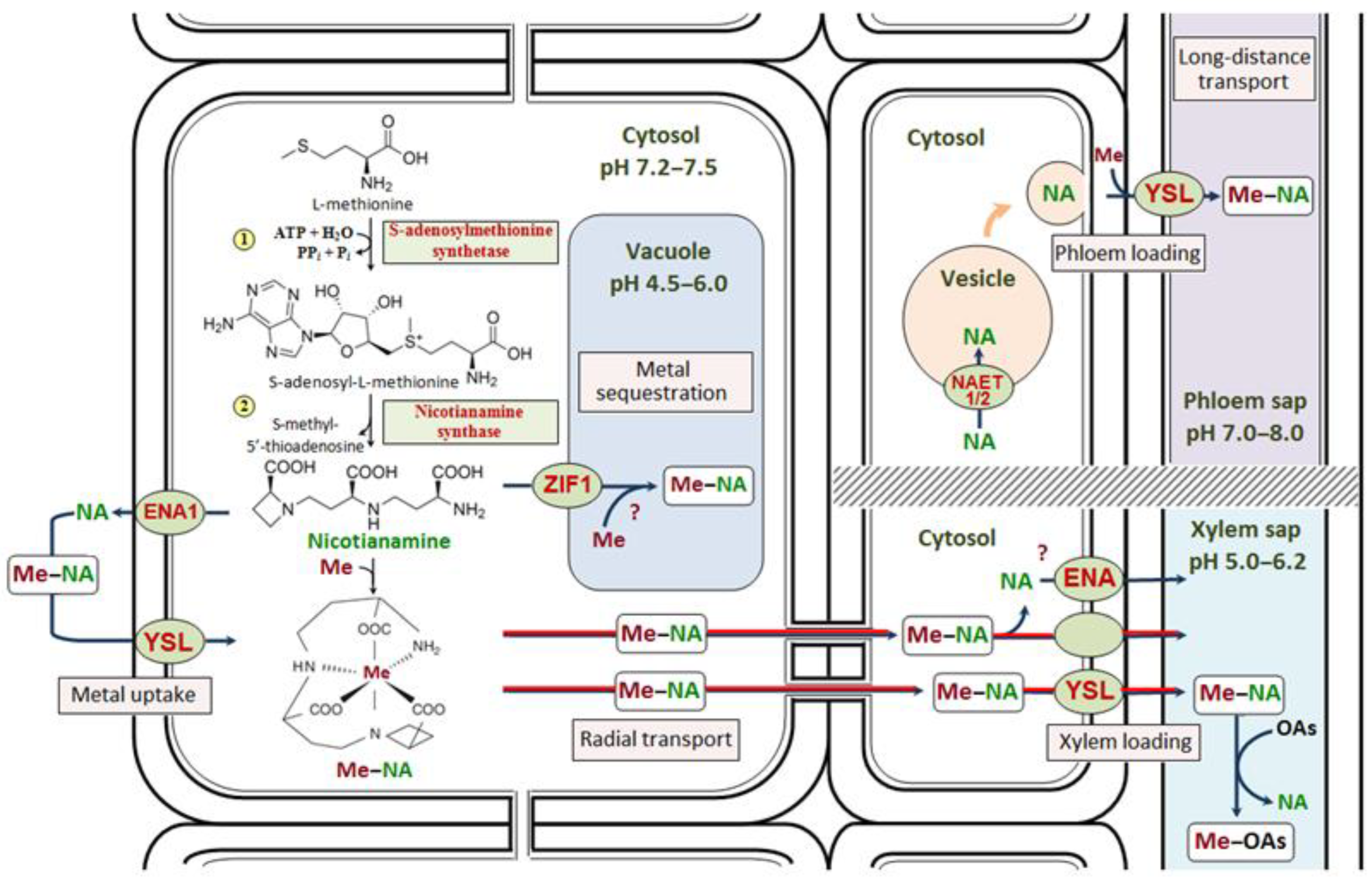
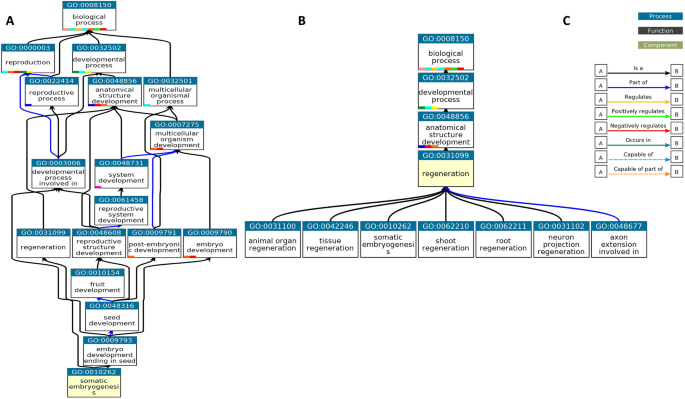
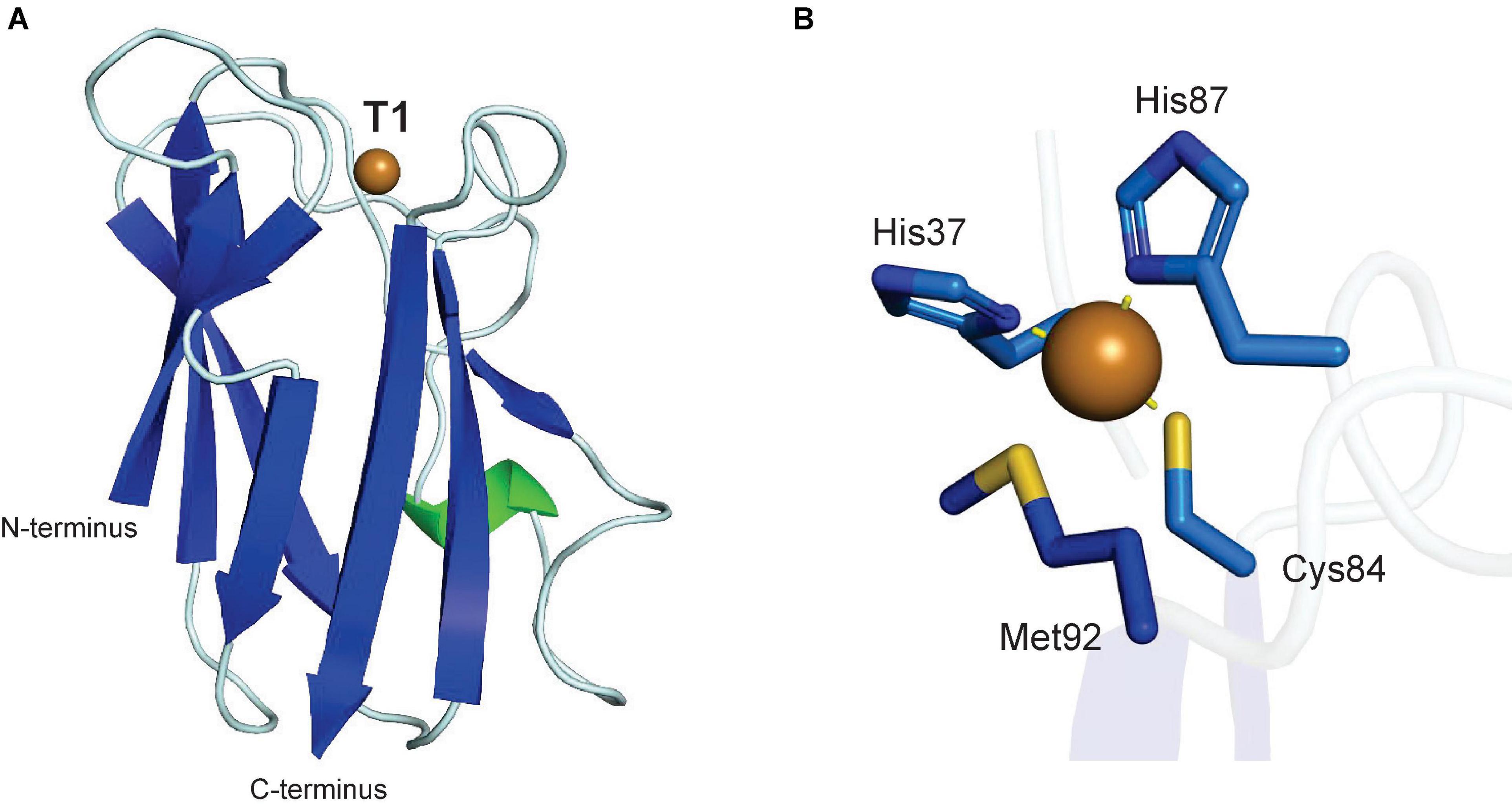
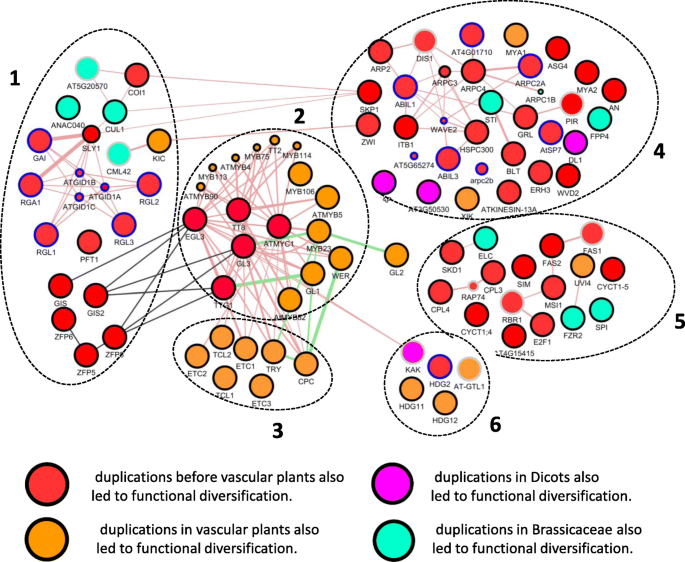
The evolution of gene regulatory networks controlling Arabidopsis
Background The variation in structure and function of gene regulatory networks (GRNs) participating in organisms development is a key for understanding species-specific evolutionary strategies. Even the tiniest modification of developmental GRN might result in a substantial change of a complex morphogenetic pattern. Great variety of trichomes and their accessibility makes them a useful model for studying the molecular processes of cell fate determination, cell cycle control and cellular morphogenesis. Nowadays, a large number of genes regulating the morphogenesis of A. thaliana trichomes are described. Here we aimed at a study the evolution of the GRN defining the trichome formation, and evaluation its importance in other developmental processes. Results In study of the evolution of trichomes formation GRN we combined classical phylogenetic analysis with information on the GRN topology and composition in major plants taxa. This approach allowed us to estimate both times of evolutionary emergence of the GRN components which are mainly proteins, and the relative rate of their molecular evolution. Various simplifications of protein structure (based on the position of amino acid residues in protein globula, secondary structure type, and structural disorder) allowed us to demonstrate the evolutionary associations between changes in protein globules and speciations/duplications events. We discussed their potential involvement in protein-protein interactions and GRN function. Conclusions We hypothesize that the divergence and/or the specialization of the trichome-forming GRN is linked to the emergence of plant taxa. Information about the structural targets of the protein evolution in the GRN may predict switching points in gene networks functioning in course of evolution. We also propose a list of candidate genes responsible for the development of trichomes in a wide range of plant species.

Regulation of transcription in plants: mechanisms controlling developmental switches

Identification of Regulatory Networks and Hub Genes Controlling Nitrogen Uptake in Tea Plants [Camellia sinensis (L.) O. Kuntze]

Identification of Conserved Gene-Regulatory Networks that Integrate Environmental Sensing and Growth in the Root Cambium - ScienceDirect

AGENT: the Arabidopsis Gene Regulatory Network Tool for Exploring and Analyzing GRNs
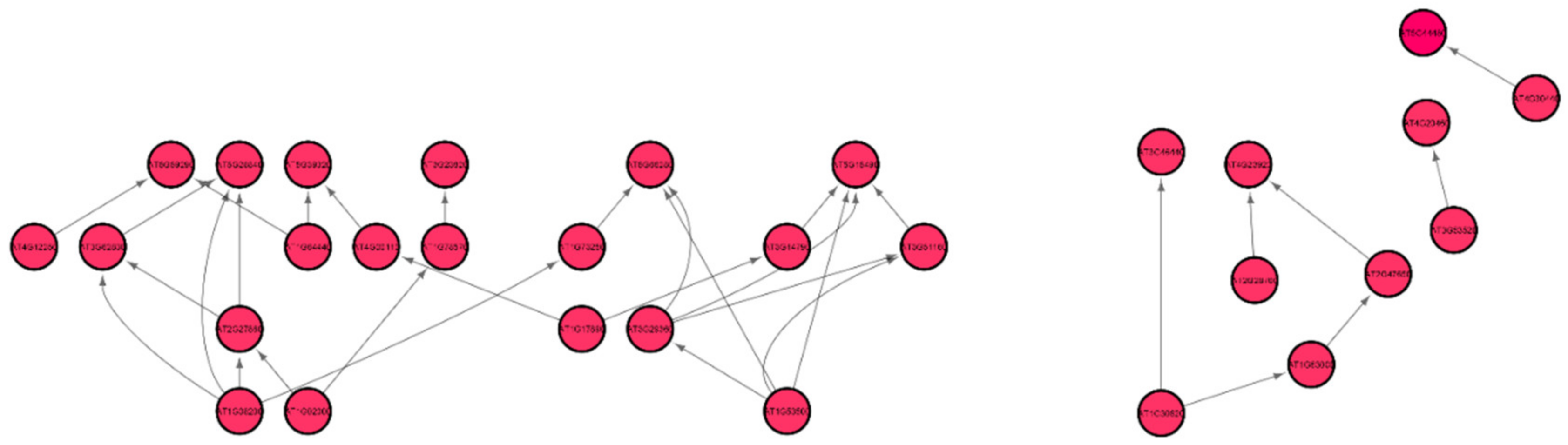
Genes, Free Full-Text
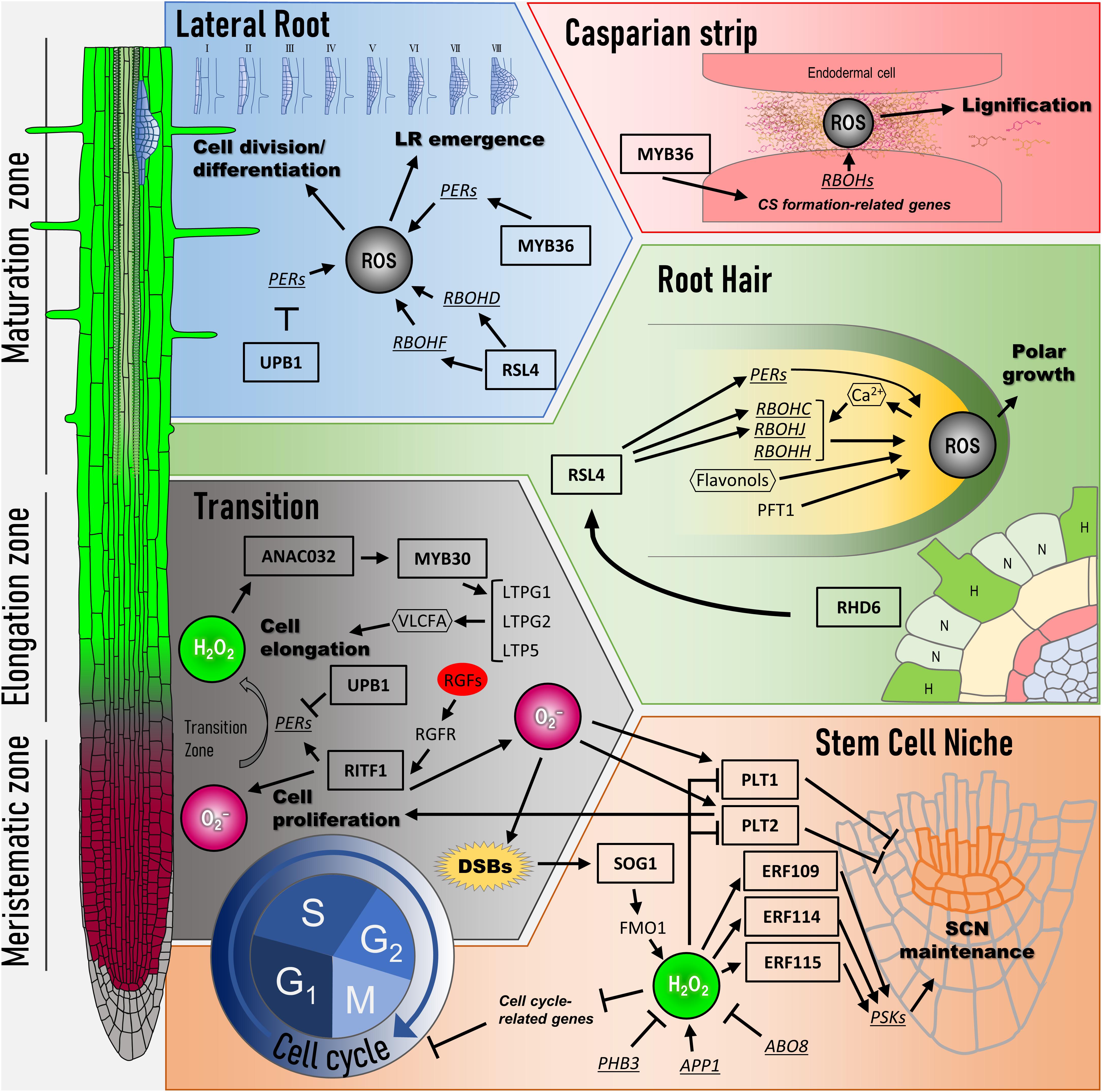
Frontiers Reactive Oxygen Species Link Gene Regulatory Networks During Arabidopsis Root Development

Gene networks controlling Arabidopsis thaliana flower development - Ó'Maoiléidigh - 2014 - New Phytologist - Wiley Online Library

Transcriptional control of photosynthetic capacity: conservation and divergence from Arabidopsis to rice - Wang - 2017 - New Phytologist - Wiley Online Library

Reconstruction of a gene regulatory network of the induced systemic resistance defense response in Arabidopsis using boolean networks, BMC Bioinformatics

Gene networks controlling the initiation of flower development: Trends in Genetics

Frontiers System Principles Governing the Organization, Architecture, Dynamics, and Evolution of Gene Regulatory Networks
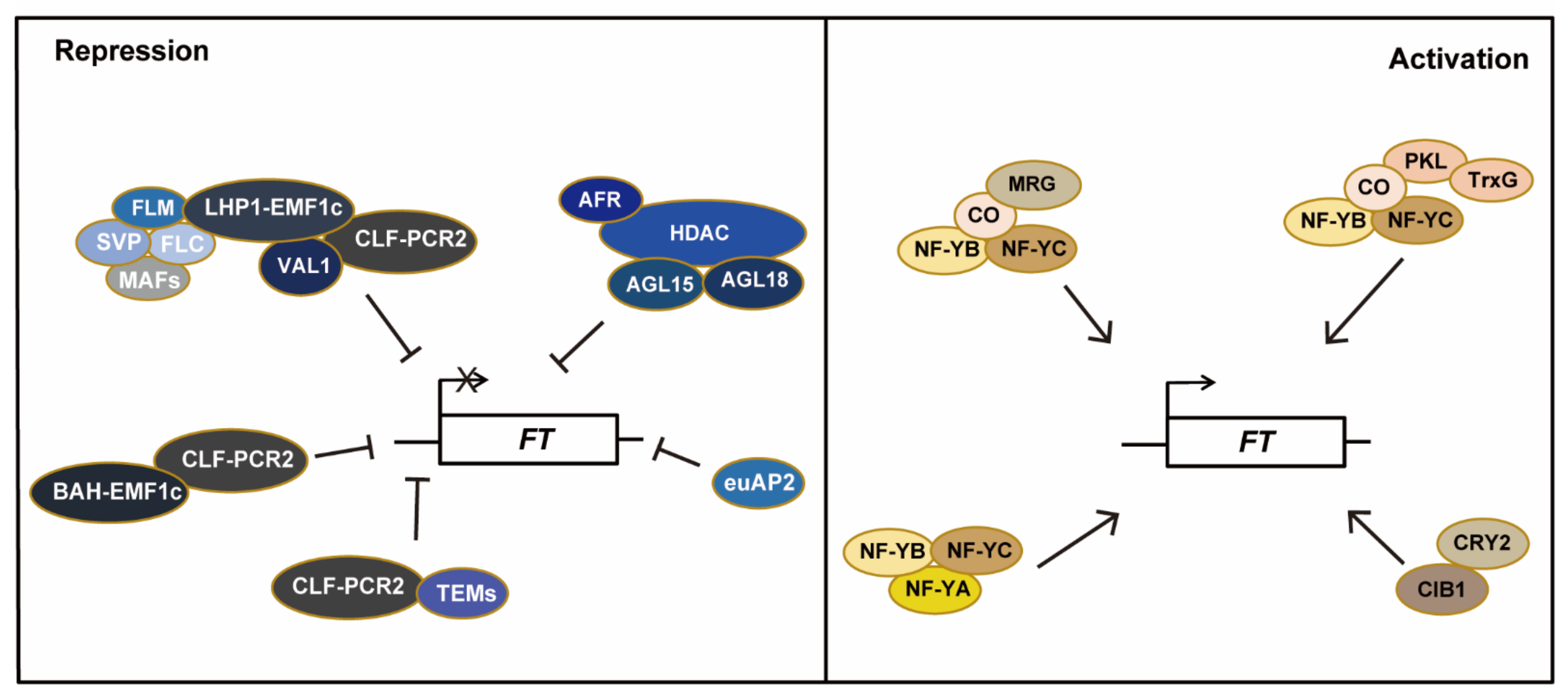
IJMS, Free Full-Text